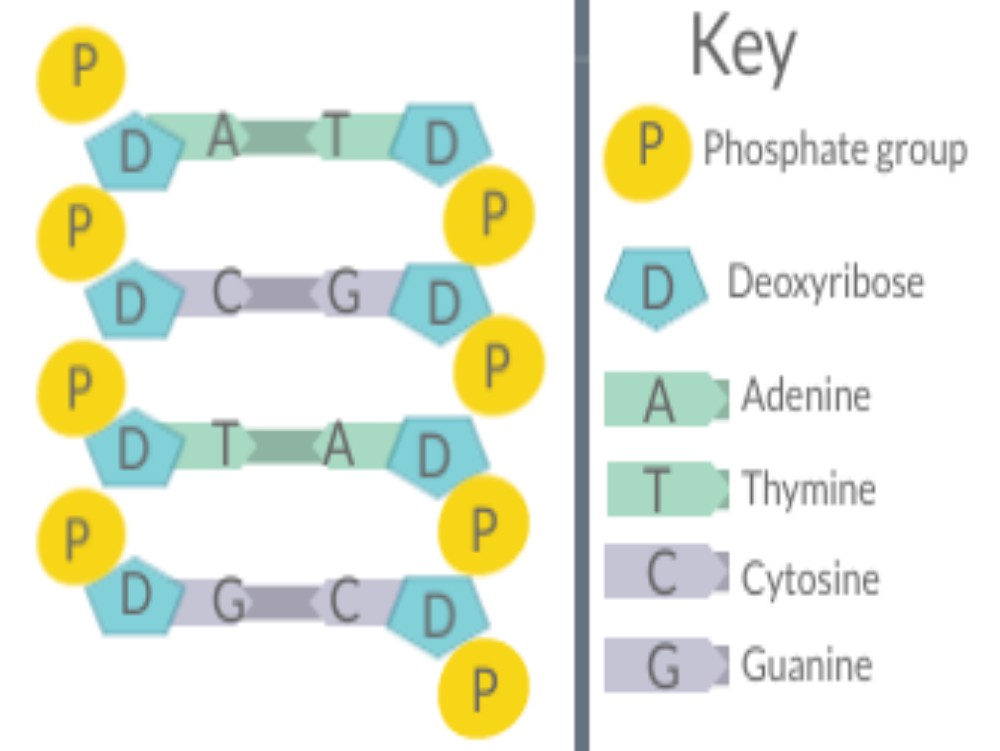
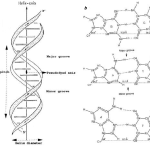
The rules of base pairing (or nucleotide pairing) are:
This is consistent with there not being enough space (20 Å) for two purines to fit within the helix and too much space for two pyrimidines to get close enough to each other to form hydrogen bonds between them.
But why not A with C and G with T?
The answer: only with A & T and with C & G are there opportunities to establish hydrogen bonds (shown here as dotted lines) between them (two between A & T;
three between C & G). The ability to form hydrogen bonds makes the base pairs more stable structurally.
These base pair relationships are often called Chargaff’s rules of DNA base pairing, named after the Columbia University scientists who observed that there are equal molar concentration of A & T, as well as G & C in most DNA molecules.
The rules of base pairing tell us that if we can “read” the sequence of nucleotides on one strand of DNA, we can immediately deduce the complementary sequence on the other strand.
The rules of base pairing explain the phenomenon that whatever the amount of adenine (A) in the DNA of an organism, the amount of thymine (T) is the same (Chargaff’s rule). Similarly, whatever the amount of guanine (G), the amount of cytosine (C) is the same.
The C+G : A+T ratio varies from organism to organism among the prokaryotes), but within (particularly the limits of experimental error, A = T and C = G.
| Relative Proportions (%) of Bases in DNA | ||||
| Organism | A | T | G | C |
| Human | 30.9 | 29.4 | 19.9 | 19.8 |
| Chicken | 28.8 | 29.2 | 20.5 | 21.5 |
| Grasshopper | 29.3 | 29.3 | 20.5 | 20.7 |
| Sea Urchin | 32.8 | 32.1 | 17.7 | 17.3 |
| Wheat | 27.3 | 27.1 | 22.7 | 22.8 |
| Yeast | 31.3 | 32.9 | 18.7 | 17.1 |
| E. coli | 24.7 | 23.6 | 26.0 | 25.7 |