Introduction
The first isolation of what we now refer to as DNA was accomplished by Johann Friedrich Miescher circa 1870. He reported finding a weakly acidic substance of unknown function in the nuclei of human white blood cells, and named this material “nuclein”. A few years later, Miescher separated nuclein into protein and nucleic acid components. In the 1920’s nucleic acids were found to be major components of chromosomes, small gene-carrying bodies in the nuclei of complex cells. Elemental analysis of nucleic acids showed the presence of phosphorus, in addition to the usual C, H, N & O. Unlike proteins, nucleic acids contained no sulfur. Complete hydrolysis of chromosomal nucleic acids gave inorganic phosphate, 2-deoxyribose (a previously unknown sugar) and four different heterocyclic bases (shown in the following diagram). To reflect the unusual sugar component, chromosomal nucleic acids are called deoxyribonucleic acids, abbreviated DNA. Analogous nucleic acids in which the sugar component is ribose are termed ribonucleic acids, abbreviated RNA. The acidic character of the nucleic acids was attributed to the phosphoric acid moiety.
The two monocyclic bases shown here are classified as pyrimidines, and the two bicyclic bases are purines. Each has at least one N-H site at which an organic substituent may be attached. They are all polyfunctional bases, and may exist in tautomeric forms.
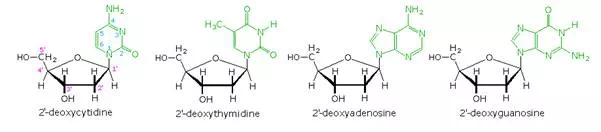
Base-catalyzed hydrolysis of DNA gave four nucleoside products, which proved to be N-glycosides of 2′-deoxyribose combined with the heterocyclic amines. Structures and names for these nucleosides will be displayed above by clicking on the heterocyclic base diagram. The base components are colored green, and the sugar is black. As noted in the 2′-deoxycytidine structure on the left, the numbering of the sugar carbons makes use of primed numbers to distinguish them from the heterocyclic base sites. The corresponding N-glycosides of the common sugar ribose are the building blocks of RNA, and are named adenosine, cytidine, guanosine and uridine (a thymidine analog missing the methyl group).
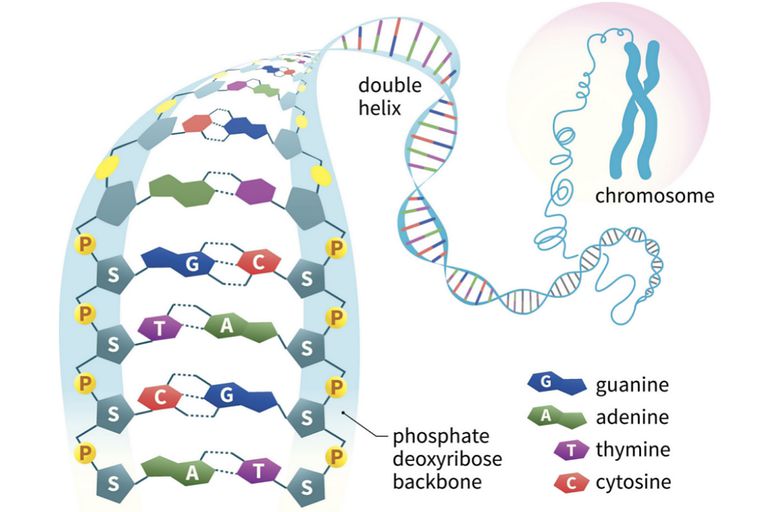
From this evidence, nucleic acids may be formulated as alternating copolymers of phosphoric acid (P) and nucleosides (N), as shown:
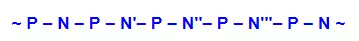
At first the four nucleosides, distinguished by prime marks in this crude formula, were assumed to be present in equal amounts, resulting in a uniform structure, such as that of starch. However, a compound of this kind, presumably common to all organisms, was considered too simple to hold the hereditary information known to reside in the chromosomes. This view was challenged in 1944, when Oswald Avery and colleagues demonstrated that bacterial DNA was likely the genetic agent that carried information from one organism to another in a process called “transformation”. He concluded that “nucleic acids must be regarded as possessing biological specificity, the chemical basis of which is as yet undetermined.” Despite this finding, many scientists continued to believe that chromosomal proteins, which differ across species, between individuals, and even within a given organism, were the locus of an organism’s genetic information.
It should be noted that single celled organisms like bacteria do not have a well-defined nucleus. Instead, their single chromosome is associated with specific proteins in a region called a “nucleoid”. Nevertheless, the DNA from bacteria has the same composition and general structure as that from multicellular organisms, including human beings.
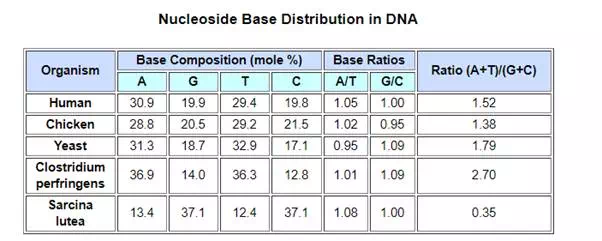
Views about the role of DNA in inheritance changed in the late 1940’s and early 1950’s. By conducting a careful analysis of DNA from many sources, Erwin Chargaff found its composition to be species specific. In addition, he found that the amount of adenine (A) always equaled the amount of thymine (T), and the amount of guanine (G) always equaled the amount of cytosine (C), regardless of the DNA source. As set forth in the following table, the ratio of (A+T) to (C+G) varied from 2.70 to 0.35. The last two organisms are bacteria.
In a second critical study, Alfred Hershey and Martha Chase showed that when a bacterium is infected and genetically transformed by a virus, at least 80% of the viral DNA enters the bacterial cell and at least 80% of the viral protein remains outside. Together with the Chargaff findings this work established DNA as the repository of the unique genetic characteristics of an organism.
The Chemical Nature of DNA
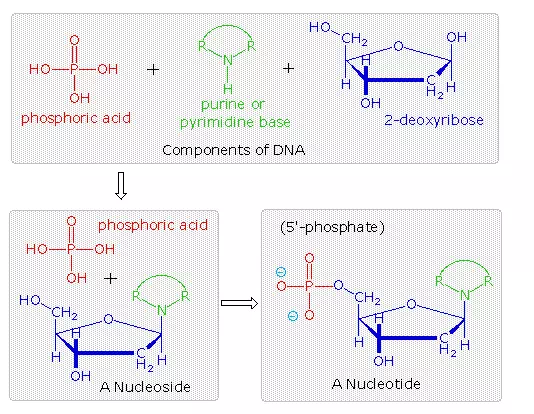
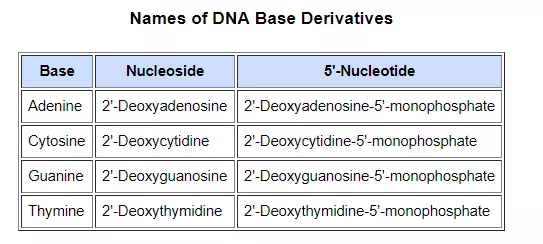
The polymeric structure of DNA may be described in terms of monomeric units of increasing complexity. In the top shaded box of the following illustration, the three relatively simple components mentioned earlier are shown. Below that on the left , formulas for phosphoric acid and a nucleoside are drawn. Condensation polymerization of these leads to the DNA formulation outlined above. Finally, a 5′- monophosphate ester, called a nucleotide may be drawn as a single monomer unit, shown in the shaded box to the right. Since a monophosphate ester of this kind is a strong acid (pKa of 1.0), it will be fully ionized at the usual physiological pH (ca.7.4). Names for these DNA components are given in the table to the right of the diagram. Isomeric 3′-monophospate nucleotides are also known, and both isomers are found in cells. They may be obtained by selective hydrolysis of DNA through the action of nuclease enzymes. Anhydride-like di- and tri-phosphate nucleotides have been identified as important energy carriers in biochemical reactions, the most common being ATP (adenosine 5′-triphosphate).
A complete structural representation of a segment of the DNA polymer formed from 5′-nucleotides may be viewed by clicking on the above diagram. Several important characteristics of this formula should be noted.
• First, the remaining P-OH function is quite acidic and is completely ionized in biological systems.
• Second, the polymer chain is structurally directed. One end (5′) is different from the other (3′).
• Third, although this appears to be a relatively simple polymer, the possible permutations of the four nucleosides in the chain become very large as the chain lengthens.
• Fourth, the DNA polymer is much larger than originally believed. Molecular weights for the DNA from multicellular organisms are commonly 109 or greater.
Information is stored or encoded in the DNA polymer by the pattern in which the four nucleotides are arranged. To access this information the pattern must be “read” in a linear fashion, just as a bar code is read at a supermarket checkout. Because living organisms are extremely complex, a correspondingly large amount of information related to this complexity must be stored in the DNA. Consequently, the DNA itself must be very large, as noted above. Even the single DNA molecule from an E. coli bacterium is found to have roughly a million nucleotide units in a polymer strand, and would reach a millimeter in length if stretched out. The nuclei of multicellular organisms incorporate chromosomes, which are composed of DNA combined with nuclear proteins called histones. The fruit fly has 8 chromosomes, humans have 46 and dogs 78 (note that the amount of DNA in a cell’s nucleus does not correlate with the number of chromosomes). The DNA from the smallest human chromosome is over ten times larger than E. coli DNA, and it has been estimated that the total DNA in a human cell would extend to 2 meters in length if unraveled. Since the nucleus is only about 5μm in diameter, the chromosomal DNA must be packed tightly to fit in that small volume.
In addition to its role as a stable informational library, chromosomal DNA must be structured or organized in such a way that the chemical machinery of the cell will have easy access to that information, in order to make important molecules such as polypeptides. Furthermore, accurate copies of the DNA code must be created as cells divide, with the replicated DNA molecules passed on to subsequent cell generations, as well as to progeny of the organism. The nature of this DNA organization, or secondary structure, will be discussed in a later section.
RNA, a Different Nucleic Acid
The high molecular weight nucleic acid, DNA, is found chiefly in the nuclei of complex cells, known as eucaryotic cells, or in the nucleoid regions of procaryotic cells, such as bacteria. It is often associated with proteins that help to pack it in a usable fashion.
In contrast, a lower molecular weight, but much more abundant nucleic acid, RNA, is distributed throughout the cell, most commonly in small numerous organelles called ribosomes. Three kinds of RNA are identified, the largest subgroup (85 to 90%) being ribosomal RNA, rRNA, the major component of ribosomes, together with proteins. The size of rRNA molecules varies, but is generally less than a thousandth the size of DNA. The other forms of RNA are messenger RNA , mRNA, and transfer RNA , tRNA. Both have a more transient existence and are smaller than rRNA.
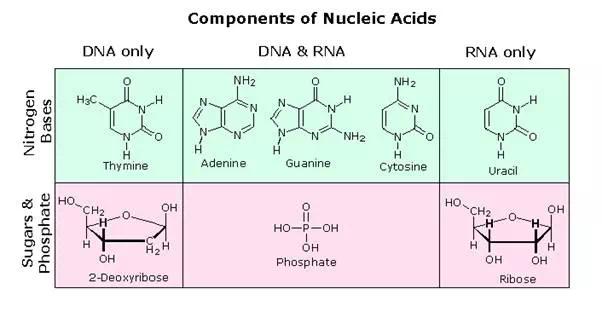
All these RNA’s have similar constitutions, and differ from DNA in two important respects. As shown in the following diagram, the sugar component of RNA is ribose, and the pyrimidine base uracil replaces the thymine base of DNA. The RNA’s play a vital role in the transfer of information (transcription) from the DNA library to the protein factories called ribosomes, and in the interpretation of that information (translation) for the synthesis of specific polypeptides. These functions will be described later.
A complete structural representation of a segment of the RNA polymer formed from 5′-nucleotides may be viewed by clicking on the above diagram
The Secondary Structure of DNA
In the early 1950’s the primary structure of DNA was well established, but a firm understanding of its secondary structure was lacking. Indeed, the situation was similar to that occupied by the proteins a decade earlier, before the alpha helix and pleated sheet structures were proposed by Linus Pauling. Many researchers grappled with this problem, and it was generally conceded that the molar equivalences of base pairs (A & T and C & G) discovered by Chargaff would be an important factor. Rosalind Franklin, working at King’s College, London, obtained X-ray diffraction evidence that suggested a long helical structure of uniform thickness. Francis Crick and James Watson, at Cambridge University, considered hydrogen bonded base pairing interactions, and arrived at a double stranded helical model that satisfied most of the known facts, and has been confirmed by subsequent findings.
Base Pairing
Careful examination of the purine and pyrimidine base components of the nucleotides reveals that three of them could exist as hydroxy pyrimidine or purine tautomers, having an aromatic heterocyclic ring. Despite the added stabilization of an aromatic ring, these compounds prefer to adopt amide-like structures. These options are shown in the following diagram, with the more stable tautomer drawn in blue.
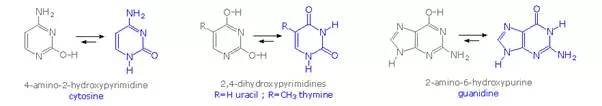
A simple model for this tautomerism is provided by 2-hydroxypyridine. As shown on the left below, a compound having this structure might be expected to have phenol-like characteristics, such as an acidic hydroxyl group. However, the boiling point of the actual substance is 100º C greater than phenol and its acidity is 100 times less than expected (pKa = 11.7). These differences agree with the 2-pyridone tautomer, the stable form of the zwitterionic internal salt. Further evidence supporting this assignment will be displayed by clicking on the diagram.
Note that this tautomerism reverses the hydrogen bonding behavior of the nitrogen and oxygen functions (the N-H group of the pyridone becomes a hydrogen bond donor and the carbonyl oxygen an acceptor).
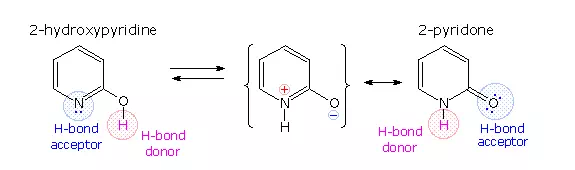
The additional evidence for the pyridone tautomer, that appears above by clicking on the diagram, consists of infrared and carbon nmr absorptions associated with and characteristic of the amide group. The data for 2-pyridone is given on the left. Similar data for the N-methyl derivative, which cannot tautomerize to a pyridine derivative, is presented on the right.
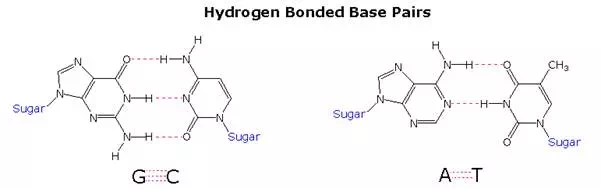
Once they had identified the favored base tautomers in the nucleosides, Watson and Crick were able to propose a complementary pairing, via hydrogen bonding, of guanosine (G) with cytidine (C) and adenosine (A) with thymidine (T). This pairing, which is shown in the following diagram, explained Chargaff’s findings beautifully, and led them to suggest a double helix structure for DNA.
Before viewing this double helix structure itself, it is instructive to examine the base pairing interactions in greater detail. The G#C association involves three hydrogen bonds (colored pink), and is therefore stronger than the two-hydrogen bond association of A#T. These base pairings might appear to be arbitrary, but other possibilities suffer destabilizing steric or electronic interactions. By clicking on the diagram two such alternative couplings will be shown. The C#T pairing on the left suffers from carbonyl dipole repulsion, as well as steric crowding of the oxygens. The G#A pairing on the right is also destabilized by steric crowding (circled hydrogens).
A simple mnemonic device for remembering which bases are paired comes from the line construction of the capital letters used to identify the bases. A and T are made up of intersecting straight lines. In contrast, C and G are largely composed of curved lines. The RNA base uracil corresponds to thymine, since U follows T in the alphabet.