In the beginning, before doing any wet lab work, design the primers complementary to the know DNA regions using the computational software.
The detail information of how to design primer is given into the article: PCR primer design guidelines or one amazing book contains all the information on primer designing. Take a look inside this amazing Springer book, I had learned primer designing from this book: PCR Primer Design
Now extract the DNA using any of the DNA extraction protocols given above.
Use 5μg DNA for restriction digestion at 37°C for 12 hours. Heat the sample for 65°C for 20 minutes to inactivate the enzyme.
The reaction preparation for the restriction digestion is given into the table below,
Restriction digestion reaction preparation:
| Reagent | Concentration |
| gDNA | 5μl (Total 5μg) |
| RE buffer (5X)* | 4μl |
| REase (restriction endonuclease) (1 unit) | 2μl |
| D/W | 9μl |
| Final volume | 20μl |
*If the concentration of RE buffer is 10X use only 2μl of buffer for the reaction.
After that, extract again the DNA using the phenol-chloroform method.
Precipitate DNA with ethanol and if precipitates are less, add a pinch of sodium acetate. Now dissolve the DNA pellets into the TE buffer.
Or
You can directly use the spin column DNA cleaning kit for cleaning the DNA sample.
Perform agarose gel electrophoresis for gDNA to know the digestion (whether the DNA is digested or not)
Use 100ng to 1 μg DNA for the ligation reaction. You can do self-ligation ( for that use 0.1- 0.2 μg DNA). The ideal reaction for the ligation assay is given into the table below,
Ligation reaction preparation:
| Reagent | Quantity |
| Template DNA | 100ng to 500ng |
| Ligase enzyme | 5 μl ( 1 unit or 1μl for 5 units) |
| Ligation buffer | 10μl |
| ATP (optional) | 10μl (10mM) |
| DD/W | Final volume 100ml |
For the ligation assay, Bacteriophage T4 ligase can be used. This bacterial ligase it the best for all kind of ligation reactions. Now incubate the ligation reaction mixture for 16°C overnight (the highest activity of ligase reported at low temperature).
With the help of the phenol-chloroform DNA extraction protocol or by the spin column DNA preparation kit, extract the DNA. If PCI is practised, precipitate the DNA with the ethanol and dissolve it in a TE buffer.
After that, quantify the DNA and use it for the PCR reaction.
1. Know the secrets of PCR reaction: PCR reaction: Ten secrets that nobody tells you
The concentration of different components used into the PCR reaction for inverse PCR is given into the table below,
PCR reaction preparation:
| Component | Stock | Concentration | Quantity |
| dNTPs | 200µM | 200µM each | 4µL |
| PCR reaction bufferWith MgCl2 | 10X | 2X | 5µL |
| Taq DNA polymerase | 5U | 1U | 0.5µL |
| Forward primer | 100pM | 200pM | 2µL |
| Reverse primer | 100pM | 200pM | 2µL |
| Ligated DNA | 250 ng | 500 ng | 2µL |
| Water | 9.5µL | ||
| Total | ————————– | 25µL |
Now our PCR reaction is ready, before doing the PCR reaction preparation if you don’t have knowledge about what precautions should be taken while preparing the PCR reaction, please read this article first: 10 tips on how to do PCR
Place the PCR tubes into the PCR machine and set the protocol as given into the table below,
PCR cycle conditions:
| PCR Steps | Initial Denaturation | Denaturation | Annealing | Extension | Final extension |
| Temperature | 90 ̊C-95 ̊C | 90 ̊C-95 ̊C | 55 ̊C-60 ̊C | 72 ̊C | 72 ̊C |
| Time | 5min | 50 sec | 50sec | 2min | 7 min |
| ——————– | ——————- | 25-30 cycles | ————— | ——————— |
After the completion of the PCR, confirm the amplicons using the agarose gel electrophoresis.
Send the sample for DNA sequencing. We already have the primer information of the known DNA sequence and based on that the sequencing is performed for the unknown DNA which is now in between the know DNA regions.
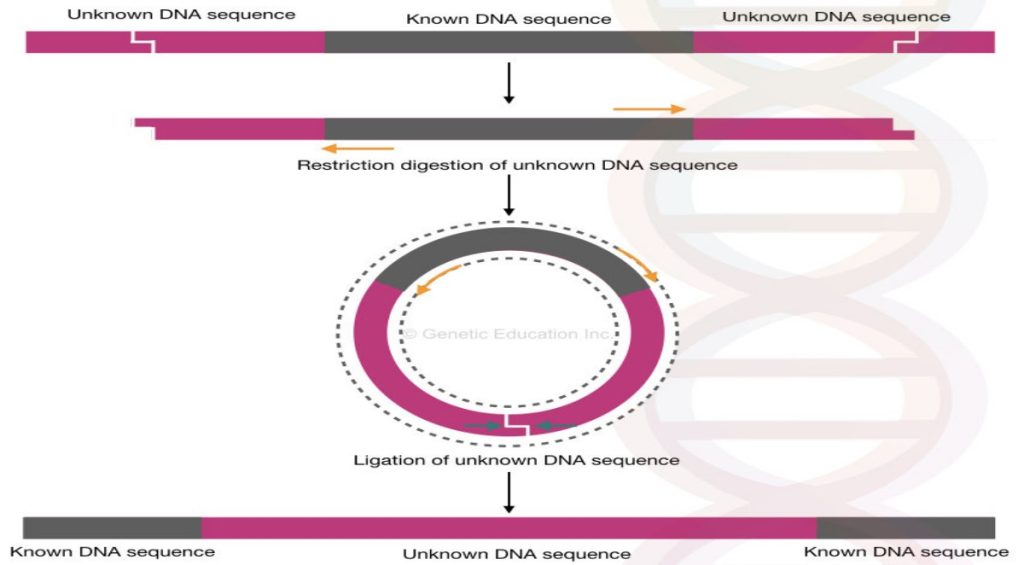
The entire mechanism of inverse PCR is shown in the figure below,

The inverse PCR is something very different than the other PCR modifications, why?
Because inverse PCR facilitates the identification of an unknown region of DNA while the conventional PCR can only be used for amplification and identification of known DNA regions or known mutations.
Now take a look at some of the application of inverse PCR:
Applications of inverse PCR:
Identification of unknown flanking regions. For example, the identification and investigation of promoter and enhancer regions of DNA upstream or downstream to the exon region can be possible by using the inverse PCR.
It is further useful in the identification of unknown mutations such as gene rearrangements, gene fusion, oncogenic gene arrangement on a chromosome.
Insertion of viral gene segments or plasmid is also investigated using the inverse PCR method.
The inverse PCR is the first choice for transposable element studies, identification and characterization.
One of the important application of the inverse PCR is in the site-directed mutagenesis. Read our site-directed mutagenesis article: Site-Directed Mutagenesis: Methods and Applications.
You can also read our series of articles on transposons:
Limitations of inverse PCR:
Although inverse PCR is a very novel method, still it is a time-consuming and tedious process. Restriction digestion and ligation assay take more time as compared to the conventional PCR.
Due to this reason, it cannot be used into the routine genetic diagnostic labs.
The technique depends on so many enzymatic steps, hence the chance of reaction failure is high.
The cost of the overall experiment is higher than the conventional PCR.
Conclusion:
The inverse PCR method is different from other PCR techniques. The method is apparently more useful in a plasmid, bacterial and plant studies in which it is applicable in unknown gene segment detection.