Radiation hybrid mapping is a genetic technique that was originally developed for constructing long-range maps of mammalian chromosomes . It is based on a statistical method to determine not only the distances between deoxyribonucleic acid (DNA) markers but also their order on the chromosomes. DNA markers are short, repetitive DNA sequences, most often located in noncoding regions of the genome , that have proven extremely valuable for localizing human disease genes in the genome.

Theory and Application
In radiation hybrid mapping, human chromosomes are separated from one another and broken into several fragments using high doses of X rays. Similar to the underlying principle of mapping genes by linkage analysis based on recombination events, the farther apart two DNA markers are on a chromosome, the more likely a given dose of X rays will break the chromosome between them and thus place the two markers on two different chromosomal fragments. The order of markers on a chromosome can be determined by estimating the frequency of breakage that, in turn, depends on the distance between the markers. This technique has been used to construct whole-genome radiation hybrid maps.
Technique
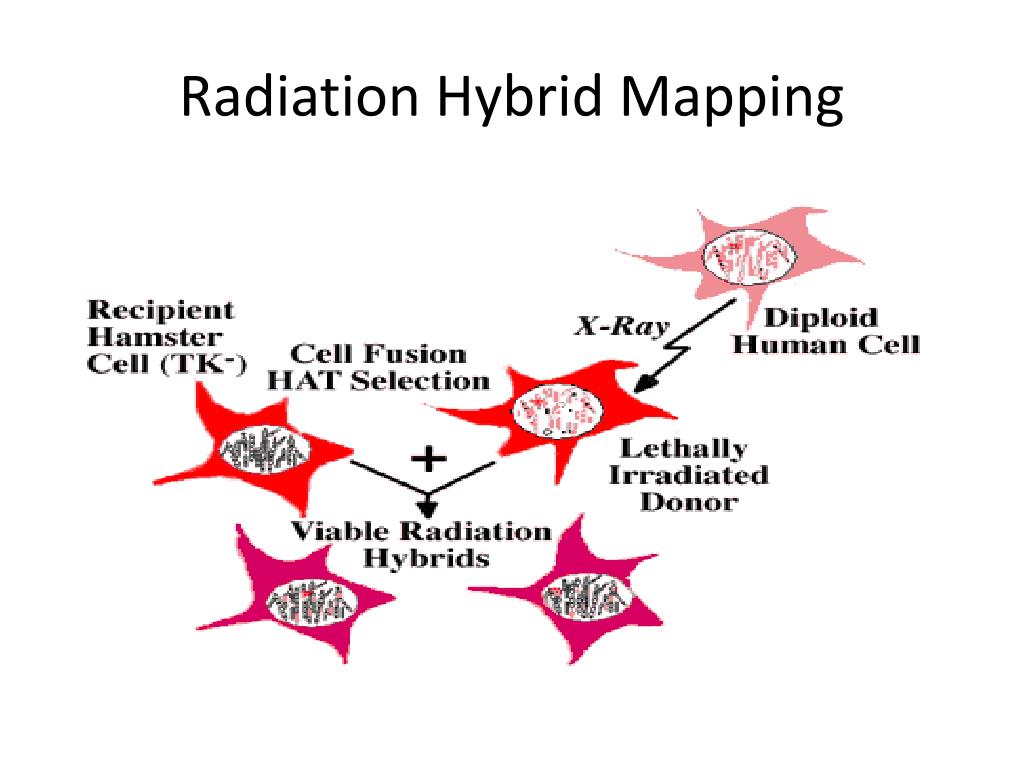
A rodent-human somatic cell hybrid (“artificial” cells with both rodent and human genetic material), which contains a single copy of the human chromosome

Radiation hybrid mapping process.
of interest, is X-irradiated. This breaks the chromosome into several pieces, which are subsequently integrated into the rodent chromosomes. In addition, the dosage of radiation is sufficient to kill the somatic cell hybrid or donor cells, which are then rescued by fusing them with nonirradiated rodent recipient cells. The latter, however, lack an important enzyme and are also killed when grown in a specific medium. Therefore, the only cells that can survive the procedure are donor-recipient hybrids that have acquired a rodent gene for the essential enzyme from the irradiated rodent-human cell line (see Figure above).
From these donor-recipient hybrids, clones can be isolated and tested for the presence or absence of DNA markers on the human chromosome of interest, and the frequencies with which markers were retained in each clone can be calculated. This process is complicated by the fact that hybrids may contain more than one DNA fragment. For example, two markers retained in one hybrid may result from retention of the two markers on separate fragments or from no break between the markers. However, the frequency of breakage, theta, can be estimated using statistical methods, and a lod score (logarithm of the likelihood ratio for linkage) can be calculated to identify significantly linked marker pairs.