INTRODUCTION
Understanding the functions of microbial cell surfaces requires knowledge of their structural and physical properties. Electron microscopy has long been recognized as a key technique in microbiology to elucidate cell surface ultrastructure . An exciting achievement has been the development of cryotechniques which allow high-resolution imaging of cell structures in conditions close to the native state. Yet direct observation in aqueous solution remained impossible.
Because of the small size of microorganisms, the physical properties of their surfaces have been difficult to study. Quantitative and qualitative information on physical properties can be obtained by electron microscopy techniques, X-ray photoelectron spectroscopy, infrared spectroscopy, contact angle, and electrophoretic mobility measurements (27). Most of these methods, however, require cell manipulation prior to examination (staining and drying), which may seriously compromise the validity of the analysis. Furthermore, the information is generally obtained from large numbers of cells and not at the level of individual cells. Thus, there is clearly a need for new, nondestructive tools capable of probing single cell surfaces at high resolution.
During the last years, atomic force microscopy (AFM ) has been used increasingly to investigate microbial surfaces at high resolution. The technique provides three-dimensional images of the surface ultrastructure with molecular resolution, in real time, under physiological conditions, and with minimal sample preparation. AFM is more than a surface-imaging tool in that force measurements can be used to probe the physical properties of the specimen, such as molecular interactions, surface hydrophobicity, surface charges, and mechanical properties. These measurements provide new insight into the structure-function relationships of microbial surfaces.
This minireview provides a survey of the various applications offered by AFM in microbiology. Before these aspects are covered, the instrumentation and methodologies are presented, and the limitations commonly encountered with microbiological specimens are discussed.
INSTRUMENTATION
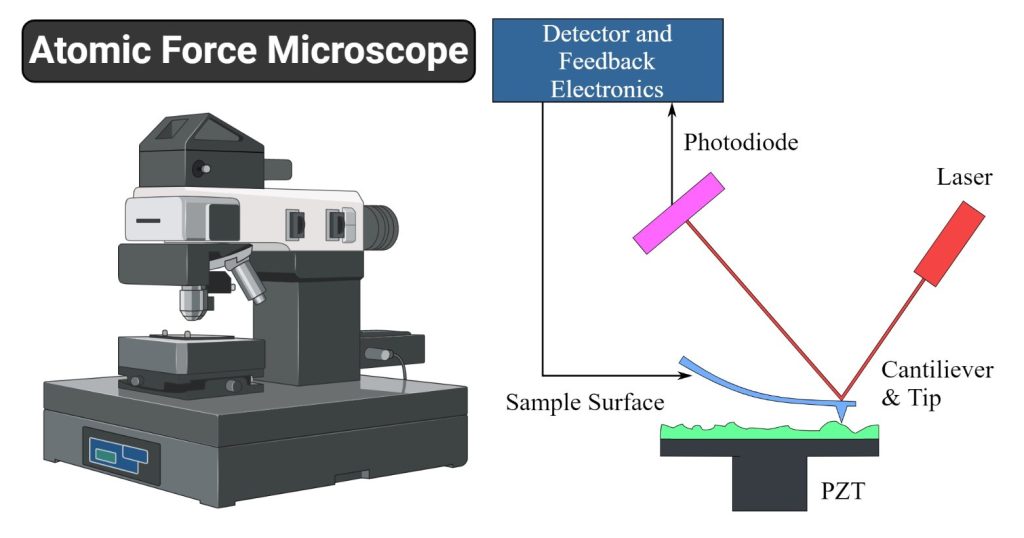
Basic elements.AFM imaging is performed not by means of an incident beam as in other classical microscopies, but by sensing the force between a very sharp probe and the sample surface . Thus, an AFM image is generated by recording the force changes as the probe (or sample) is scanned in the x and y directions. The sample is mounted on a piezoelectric scanner, which ensures three-dimensional positioning with high resolution. The force is monitored by attaching the probe to a pliable cantilever, which acts as a spring, and measuring the bending or “deflection” of the cantilever. The larger the cantilever deflection, the higher the force that will be experienced by the probe. Most instruments today use an optical method to measure the cantilever deflection with high resolution; a laser beam is focused on the free end of the cantilever, and the position of the reflected beam is detected by a position-sensitive detector (photodiode). AFM cantilevers and probes are typically made of silicon or silicon nitride by microfabrication techniques.
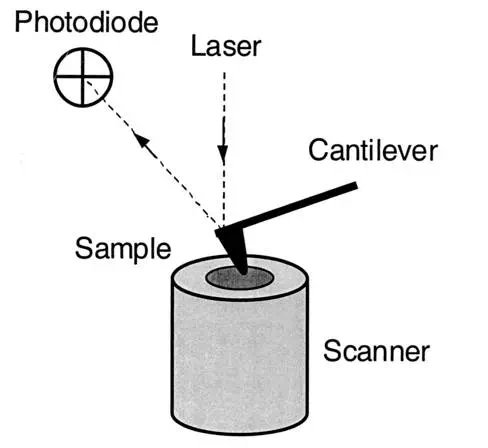
FIG. 1.
General principle of AFM.
Imaging modes.A number of AFM imaging modes are available. The most widely used imaging mode is the contact mode, in which sample topography can be measured in different ways. In the constant-height mode, one simply records the cantilever deflection while the sample is scanned horizontally, i.e., at constant height. Minimizing large deflections, thus holding the applied force to small values, is often necessary to prevent sample damage. This is achieved in the constant-deflection mode, in which the sample height is adjusted to keep the deflection of the cantilever constant by using a feedback loop. The feedback output is used to display a true “height image.” In many cases, small cantilever deflections do occur because the feedback loop is not perfect, and the resulting error signal can be used to generate a so-called “deflection image.” The height image provides quantitative height measurements, allowing accurate measurement of surface roughness, the height of surface features, or the thickness of biological layers. The deflection image does not reflect true height variations, but because the frequency response is much higher, it is more sensitive to fine surface details than the height signal.
Several other AFM imaging modes have been developed, such as the tapping mode, in which the probe is excited externally and the amplitude and phase of the cantilever are monitored near the resonance frequency of the cantilever. Although the use of tapping mode AFM in microbiology has been limited so far, it has a great potential for imaging surface topography with minimal applied forces.
Force measurements.Measuring the force acting between the AFM probe and the sample, by means of force-distance curves, is important in defining the imaging force and thus in optimizing the image resolution. Furthermore, as discussed below, AFM force measurements can be used to probe the sample’s physical properties. Force-distance curves are recorded by monitoring, at a given x–y location, the cantilever deflection as a function of the vertical displacement of the piezoelectric scanner. A raw curve is a plot of the photodiode voltage versus the scanner position. By using appropriate corrections, this can be converted into a force-versus-separation distance curve.
The different parts of a force-distance curve can provide a wealth of information. At large probe-sample separation distances, the force experienced by the probe is null. As the probe approaches the surface, the cantilever may bend upwards due to repulsive forces until it jumps into contact when the gradient of attractive forces exceeds the spring constant plus the gradient of repulsive forces. The approach portion of the force-distance curve can be used to measure surface forces, including van der Waals and electrostatic, solvation, hydration, and steric/bridging forces. When the force is increased in the contact region, the shape of the approach curve may provide direct information on the elasticity of the sample. When the probe is retracted from the surface, the curve often shows a hysteresis referred to as the adhesion “pull-off” force, which can be used to estimate the surface energy of solids or the binding forces between complementary biomolecules. In the presence of long, flexible molecules, an attractive force, referred to as an elongation force, may develop nonlinearly due to macromolecular stretching (for more details on the principle of force-distance curves, see reference .
LIMITATIONS
Although AFM has great potential in microbiology, it is important to realize that accurate data collection is often delicate due to the several limitations and difficulties associated with the technique. One of the most crucial aspects is sample preparation. Indeed, to withstand the force exerted by the scanning probe, the sample must be well attached to an appropriate solid substrate. For reconstituted microbial layers, this is generally achieved by physical adsorption of the layers on a flat substrate, i.e., mica, glass, or silicon oxide, in the presence of appropriate electrolytes (15, 30). For imaging living cells, however, immobilization by means of simple adsorption procedures is inappropriate because the contact area between cells and substrate is very small, often leading to cell detachment by the scanning probe. Therefore, several approaches have been developed to promote cell attachment.
In the air-drying method, a droplet of a concentrated cell suspension is placed on a mica or glass substrate and allowed to dry in air (1). Alternatively, covalent bonding of bacteria to silanized glass can be used. Typically, glass slides are first modified with aminosilane molecules, and the silanized slides are then reacted with a bacterial suspension containing 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide and N-hydroxysuccinimide (14).
To avoid air-drying and chemical treatments, which may cause denaturation of the surface molecules, an immobilization procedure with porous membranes has been developed (Fig. 2) (16, 22). Briefly, a concentrated cell suspension is gently sucked through an isopore polycarbonate membrane with a pore size comparable to the cell size. After the filter is cut, the lower part is carefully dried on a sheet of tissue, and the specimen is then attached to the sample holder with a small piece of adhesive tape. This approach can be used to image single bacterial, yeast, and fungal cells under aqueous conditions while minimizing denaturation of the surface molecules. However, the method essentially works for spherical cells but not for rod-shaped cells, emphasizing the need to develop other nondestructive immobilization strategies in future research.
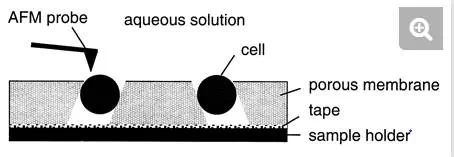
FIG. 2.
Schematic illustration of the porous membrane immobilization procedure developed for AFM imaging of living cells.
A second source of problems is related to the resolution and interpretation of the images, which depend directly on the imaging force and the probe geometry. Large forces acting between probe and sample during imaging may dramatically reduce the image resolution and cause molecular damage or displacement. Therefore, to obtain reliable, high-resolution images, microscopists must control the imaging force via force-distance curve measurements. When imaging in air, a layer of water condensation and other contamination often covers both probe and sample, forming a meniscus pulling the two together. The resulting strong attractive force, usually 10 to 100 nN, makes high-resolution imaging difficult and sometimes causes sample damage. The capillary force can be eliminated by performing the imaging in aqueous solution. By selecting appropriate buffer conditions, it is generally possible to maintain an applied force in the range of 0.1 to 0.5 nN (33). Therefore, a general recommendation for microscopists studying new microbial samples is to investigate the effect of pH and ionic strength on the image quality in order to define an optimal imaging environment. Also, because of thermal drift, it is often essential to readjust the applied force between image acquisitions.
The size and shape of the AFM probe also play an important role in determining the image contrast. At first glance, it is surprising to see that (sub)nanometer resolution is routinely acquired on reconstituted surface layers by using commercial probes with a 10- to 50-nm radius of curvature. It is thought that nanoscale protrusions or asperities extending irregularly from the probe are responsible for the high resolution. A common source of artifacts in imaging is the broadening of the surface features due to the finite size of the AFM probe. This effect is important when imaging height variations in the range of the probe’s radius of curvature, i.e., 10 to 50 nm. In this case, the sample interacts with the sides of the probe, and the resulting image will be a combination of the real sample topography and the probe geometry. Another common problem is the shadowing or multiplication of small structures produced by multiple probe effects. These are due to the presence of multiple asperities on the probe apex, which usually originate from contamination.
IMAGING SURFACE STRUCTURE
Visualizing cell surface layers at submolecular resolution.During the last decade, the unique ability of AFM to image specimens at subnanometer resolution and in aqueous solution has been intensively exploited to investigate the structure of cell surface layers made of two-dimensional protein crystals. Traditionally, X-ray crystallography has been the premier technique for atomic resolution of protein crystals, while electron crystallography examines the specimen in a more native-like environment at near-atomic resolution (35). Because it directly probes the actual surface under physiological conditions, AFM is complementary to these two techniques.
A survey of AFM studies focusing on cell surface layers is given in Table 1. S-layers of Bacillusspecies were among the first reconstituted structures to be imaged at high resolution, the images recorded in buffer solution showing oblique and square lattices that were in good agreement with transmission electron microscopy data (36). More recently, the oblique lattice of the S-layer of Bacillus stearothermophilus was visualized to a lateral resolution of about 1.5 nm (38), and nanometer lateral resolution was achieved for the S-layers of B. coagulans and B. sphaericus strains recrystallized on silanized silicon substrates (39). Interestingly, an elegant preparation method has been developed to investigate S-layers from Bacillus species in conditions relevant to their native state (49). The proteins are recrystallized on a lipid monolayer in a Langmuir-Blodgett trough, and the composite lipid/S-layer structure is then deposited on a flat substrate. Under these conditions, S-layers attach to the lipid film with their inner face, which corresponds to the orientation found in the living organism.
Several other two-dimensional protein crystals have been investigated, one of which is the hexagonally packed intermediate layer of Deinococcus radiodurans. This specimen was resolved with a lateral resolution of 1 nm (21), and the inner surface was shown to have a protruding core with a central pore exhibiting two conformations, one with and the other without a central plug (29).
The purple membrane from the archaeon Halobacterium is another well-studied example. In early work, the hexagonal symmetry of purple membranes adsorbed on mica was imaged in aqueous conditions, the lattice constant being in agreement with electron diffraction data (11). Later, conditions were established to reproducibly acquire images of purple membranes at subnanometer resolution (28). Recently, tapping mode AFM was shown to faithfully record high-resolution images of purple membranes and to have sufficient sensitivity to contour individual peptide loops without detectable deformations (25). Hence, for soft, fragile microbial samples, this imaging mode may represent a valuable alternative to contact mode, since probe-sample lateral forces are greatly reduced. Gram-negative bacteria are protected by an outer membrane in which trimeric channels, the porins, facilitate the passage of small solutes. Reconstitution of the porin OmpF crystals of Escherichia coli in the presence of lipids made it possible to visualize their structure in aqueous solution to a lateral resolution of 1 nm and a vertical resolution of 0.1 nm (42). To assess the accuracy of the AFM images, averaged surface contours were directly compared with the protein structure from X-ray analysis, excellent agreement being obtained between the subtlest features of the images and the models. Another example is aquaporins, ubiquitous membrane channels that are highly specific for water or small uncharged hydrophilic solutes and are involved in osmoregulation. For E. coli aquaporin Z, AFM images revealed two-dimensional crystals with p42(1)2 and p4 symmetry (43). Imaging both crystal types before and after cleavage of the N termini allowed the cytoplasmic surface to be identified.
AFM reveals conformational changes in relation with function.Although numerous cell surface layers have been fully characterized from the structural point of view, their specific functions often remain poorly understood. Recent data indicate that AFM has strong potential for study of the structure-function relationships of surface layers. In particular, conformational changes of individual membrane proteins can be detected and related to function. Individual pores of the hexagonally packed intermediate layers were observed to switch from one state to the other (29, 31). Along the same line, force-induced conformational changes in purple membranes were directly visualized by AFM (31); when the imaging forces were lowered from 300 to 100 pN, donut-shaped trimers reversibly transformed into structures exhibiting three protrusions. Interestingly, voltage- and pH-dependent conformational changes of the extracellular loops of OmpF porins were demonstrated (32); the observed conformational changes were correlated with closure of the channel entrance, suggesting that this is a mechanism to protect the cells from drastic changes in the environment. As discussed below, AFM force measurement is another promising method for investigating structure-function relationships.
Imaging living cells in real time and at high resolution.AFM is currently the only technique that can image the surface of a living cell at high resolution and in real time. Thus, it is complementary to electron microscopy, for which vacuum conditions are required and real-time analysis is not possible. A survey of reports dealing with living cells (no chemical fixation or dehydration)
As pointed out before (Fig. 2), mechanical trapping in porous membranes is a convenient sample preparation method to image single cells under aqueous solution without chemical pretreatment. Figure 3 shows a three-dimensional height image of a Saccharomyces cerevisiaecell trapped in a pore of the membrane. The cell surface is very smooth and shows a circular protrusion attributed to a bud scar. The main advantage of using AFM at a low resolution, such as here, is that dynamic events, such as cell growth and budding processes (18, 22) and the change in cell surface morphology resulting from treatment with external agents (e.g., enzymes and antibiotics), can be explored in real time.
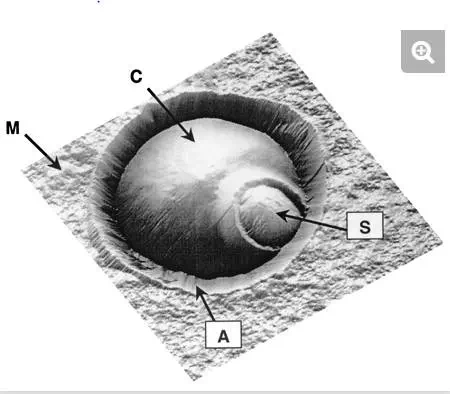
FIG. 3.
AFM height image (6 μm by 6 μm; z range, 1.5 μm), in aqueous solution, of a single Saccharomyces cerevisiae cell trapped in a porous membrane. The cell (C) is located at the center of the image, while the surrounding flat area represents the polymer membrane (M). The ∼1-μm circular protrusion is attributed to a bud scar (S). Note that the cell is surrounded by an artifactual structure (A) resulting from the contact between the AFM probe and the pore edges.
More importantly, several recent reports have indicated that AFM can resolve the surface structure of living cells at high resolution. Rodlets are well-ordered structures, known to be composed essentially of proteins and to cover the surface of the spores of many fungi, including ascomycetes, basidiomycetes, and zygomycetes (5). For the first time, these structures were visualized under aqueous solution by AFM. For example, the surface of Aspergillus oryzae spores in the dormant state displayed rodlets, 10 ± 1 nm in diameter, that were assembled in parallel to form supramolecular fascicles having different orientations (Fig. 4A) (47). Similar structures were imaged at the surface of Phanerochaete chrysosporium (16). By contrast, the yeast S. cerevisiae had a very smooth surface morphology, consistent with an outer layer of mannoproteins (Fig. 4B). In contrast to the two previous examples, the bacterium Lactococcus lactis showed a sponge-like network with small holes uniformly distributed accross the surface (Fig. 4C) (9). The observed surface structure depended on the scanning direction, indicating that the apparent surface morphology was strongly influenced by the probe-cell interaction. This example shows that while near-molecular resolution can be achieved with fungi, obtaining such resolution with bacteria remains very challenging due to sample deformation. Hence, there is a need to develop improved preparation procedures and imaging conditions for bacterial cells.
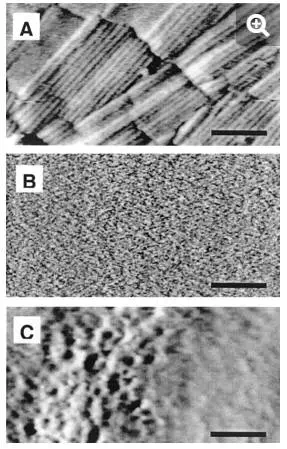
FIG. 4.
High-resolution deflection images recorded in aqueous solution for the surface of (A) Aspergillus oryzae, (B) Saccharomyces cerevisiae, and (C) Lactococcus lactis. Bars: 100 nm (A, C) and 50 nm (B). The surface of A. oryzae shows rodlet structures, thought to play different biological functions, which is in contrast to the very smooth morphology of the S. cerevisiae surface. For the bacterium L. lactis, the surface relief is strongly influenced by probe-cell interactions.
AFM has also proved useful for imaging the morphology of bacterial biofilms on solid surfaces in the hydrated state. While high-resolution information cannot be obtained due to high sample softness, direct visualization of adherent bacteria and of hydrated extracellular polymeric substances is made possible. For instance, Pseudomonas aeruginosa cells and their associated extracellular polymeric substances were observed in their hydrated forms in relation to the process of stainless steel corrosion (46). Another example is the imaging of unsaturated biofilms of Pseudomonas putida, i.e., biofilms grown in humid air (4). The surface morphology, roughness, and adhesion forces in the outer and basal cell layers of fresh and desiccated biofilms were determined. It was found that the extracellular polymeric substances formed “mesostructures” which were much larger than the discrete polymers of glycolipids and proteins that have been characterized previously on the outer surface of these gram-negative bacteria.
EXPLORING PHYSICAL PROPERTIES
Because microbial cells are so small, their physical properties have traditionally been difficult to study at the subcellular level. In recent years, AFM force measurements have made it possible to probe molecular interactions (e.g., hydrophobic and electrostatic interactions), map physicochemical properties (surface hydrophobicity), measure the stiffness of cell wall components and surface appendages, and stretch surface macromolecules. A survey of these experiments is given in Table 3.
Molecular interactions and physicochemical properties.Cellular events such as microbial adhesion, microbial aggregation, and molecular recognition play a pivotal role in the natural environment, in medicine, and in biotechnological processes. Understanding the molecular basis of these phenomena requires knowledge of the molecular interactions at the cell surface. Several approaches have been developed towards the goal of probing molecular interactions and physicochemical properties by AFM. In the so-called cell probe method, the cells are attached directly to the AFM probe, and force-distance curves are then recorded between the cell-coated probe and a solid substrate. In this way, the forces between E. coli-coated probes and solids of different surface hydrophobicity were measured (37). Both attractive forces and cell adhesion behavior were promoted by substrate surface hydrophobicity, pointing to the role of hydrophobic interactions. In the field of biomaterials, cell-coated probes were used to measure the forces between E. coli strains and polymer biomaterials (40) and substrates coated with block copolymers (41). Polymeric brush layers appeared not only to block the long-range attractive forces of interaction between bacteria and substrates but also to introduce repulsive steric effects.
The cell probe approach is also useful in biogeochemistry and geomicrobiology for studying the mineral-microbe interface. Interfacial and adhesion forces between E. coli cells and various mineral surfaces (muscovite, goethite, and graphite) were directly measured in aqueous solution (23). Attractive and repulsive interfacial forces were detected at separation ranges of up to 400 nm, the magnitude and sign depending on the ionic strength of the intervening solution and the mineral surface charge and hydrophobicity. Interaction forces between Shewanella oneidensis and goethite were measured in real time in aerobic and anaerobic solutions (24). Energy values derived from these measurements showed that the affinity between S. oneidensis and goethite increased rapidly by two- to fivefold under anaerobic conditions, in which electron transfer from bacterium to mineral is expected. It is worth noting that the cell probe method has some limitations: the treatment used to firmly attach the cells to the AFM probe may modify the cell surface macromolecular architecture, and the high lateral resolution of AFM can no longer be used.
Alternatively, force-distance curves can be recorded between an AFM probe and cells immobilized on a solid substrate. By using silicon nitride probes, the relative contributions of electrostatic and steric interactions associated with negatively charged bacterial strains were recently investigated (13). The forces measured as a function of pH and ionic strength were well represented by an electrosteric repulsion model but were much larger in magnitude and extended over longer distances than theoretically predicted. Partially removing polysaccharides from the bacterial surfaces resulted in lower repulsive forces that decayed much more rapidly.
Because most commercial AFM probes have a poorly defined surface chemistry, a valuable strategy for quantitative force measurements is to functionalize the probe surface with self-assembled monolayers terminating in specific chemical groups. As an example, the surface hydrophobicity of P. chrysosporium spores was recently mapped by recording multiple force-distance curves by using OH- (hydrophilic) and CH3– (hydrophobic) terminating probes (17). The curves always showed no adhesion, indicating that the spore surface was uniformly hydrophilic. Presumably, the nonsticky character of the fungal spore surface must play a role in determining its biological functions, namely, protection and dispersion. In the same way, AFM probes functionalized with ionizable groups could be used to characterize electrostatic interactions and cell surface charge. Hence, AFM with chemically functionalized probes appears to complement classical experimental methods used to assess microbial surface hydrophobicity and surface charge.
Finally, AFM probes can also be functionalized with biomolecules in order to measure forces between individual ligands and receptors (20). Such studies have great potential in microbiology for mapping specific recognition sites at cell surfaces and open the door to new biomedical applications.
Cell surface elasticity.How stiff is the surface of a microbial cell? This question can now be addressed on a local scale by pressing the AFM probe onto cells or isolated cell wall components. In this way, the effective compressibility of the cell wall of Magnetospirillum gryphiswaldense was determined to be about 42 mN/m (2). Furthermore, a theoretical expression was derived for the force exerted by the wall on the cantilever as a function of the depth of indentation generated by the AFM tip (3). Evidence was provided that this reaction force is a measure of the turgor pressure of the bacterium. Making use of experimental and theoretical results, the turgor pressure was determined to be in the range of 85 to 150 kPa.
Besides cell walls, surface appendages may also modulate cell surface softness. By using approach force-distance curves, significant differences in the surface softness of the fibrillated Streptococcus salivarius HB and the nonfibrillated S. salivarius HBC12 strains were demonstrated (48). In agreement with AFM and electron microscopy images, a long-range repulsion was measured on the fibrillated strain and attributed to the compression of the soft cell surface fibrils.
The elastic properties of isolated bacterial cell wall components can also be explored by using the so-called depression technique (50, 51). In this technique, a solution containing the cell wall components is placed on a hard substrate and allowed to dry. The substrate contains grooves that are narrow compared to the width or the length of the material to be investigated, so that single wall components bridging one or more grooves can be obtained. The force measurement consists of placing the probe at the midpoint of the unsuspended wall component region and increasing or decreasing the force as a linear function of time. Comparison of the cantilever deflections between specimen and hard substrate allows an accurate determination of the specimen’s elasticity. Applying this approach to the proteinaceous sheath of the archaeon Methanospirillum hungatei GP1 yielded an elastic modulus of 2 × 1010 to 4 × 1010 N/m2, indicating that the sheath could withstand an internal pressure of 400 atm, well beyond that needed for a eubacterial envelope to withstand turgor pressure. For murein sacculi of gram-negative bacteria in the hydrated state, elastic moduli of 2.5 × 107 N/m2 were measured, in excellent agreement with theoretical calculation of the elasticity of the peptidoglycan network (10).
Elasticity of surface macromolecules.The remarkable force sensitivity of AFM allows the forces associated with single molecules to be measured. When the AFM probe touches a macromolecular surface, the retraction curve often shows attractive elongation forces, i.e., adhesion peaks developing nonlinearly, reflecting the stretching of the flexible macromolecules. In the case of the hexagonally packed intermediate layer (Fig. 5), force-extension curves recorded for the inner surface showed sawtooth patterns, with six adhesion force peaks of about 300 pN (34). This behavior was well fitted with a worm-like chain model from statistical mechanics and attributed to the sequential pulling out of the protomers of the hexameric hexagonally packed intermediate protein complex. After recording the force curve, a molecular defect the size of a hexameric complex was clearly visualized by high-resolution imaging (Fig. 5A and C), indicating that an entire hexamer had been zipped out of the surface layer. This work shows that AFM has great potential for manipulating cell surface supramolecular structures and for studying their mechanical stability at the single-molecule level.
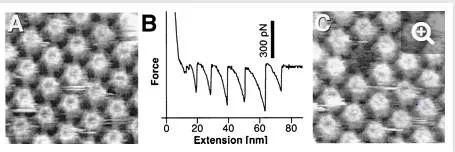
FIG. 5.
Using AFM force measurements to unzip proteins from the hexagonally packed intermediate layer (34). (A) Control AFM topograph of the inner surface of the hexagonally packed intermediate layer. (B) Force-extension curve recorded for this inner surface region, showing a sawtooth pattern with six force peaks of about 300 pN. (C) The same inner surface area imaged after recording the force curve; a molecular defect the size of a hexameric hexagonally packed intermediate protein complex has clearly been created. (Reprinted with permission of A. Engel, Universität Basel, Basel, Switzerland; copyright 1999, National Academy of Sciences.)
Similar force behaviors have been reported for whole cells. Force-extension curves recorded between living S. oneidensis bacteria and goethite showed specific signatures which were suggested to reflect the unfolding of a 150-kDa putative iron reductase (24). In another study, long macromolecules, presumably polysaccharides, were stretched at the surface of germinating spores of A. oryzae (47). Force-extension curves showed attractive elongation forces with 400 ± 100 pN adhesion peaks (Fig. 6). Elongation forces were well fitted with an extended freely jointed chain model by using fitting parameters that were consistent with the stretching of individual polyglucose molecules. It was suggested that the elastic properties of these long surface macromolecules may play a role in modulating cellular events such as cell adhesion and aggregation.
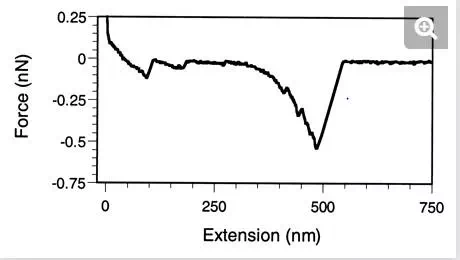
FIG. 6.
Probing the elasticity of cell surface macromolecules (47). Force-extension curve recorded on a germinating spore of A. oryzae. The elongation forces are attributed to the elastic deformation of polysaccharide chains, in agreement with the expected biochemical composition of the spore surface.
The above studies indicate that AFM force measurements will have a significant impact on future microbiological research by increasing our understanding of cell surface properties and elucidating structure-function relationships. However, acquiring reliable force data is not trivial, and several limitations and problems must be considered. First, because AFM does not provide an independent measure of the probe-sample separation distance, defining the point of contact (zero separation) may be very difficult. Second, for soft samples such as most microbial surfaces, separating the relative contributions of repulsive surface forces and sample deformation is a delicate task, and quantitative interpretation of the curves by using theoretical models may be meaningless. Third, when recording force-distance curves, attention should be paid to artifacts such as periodic oscillations in the noncontact region and offsets between the lines in the contact and noncontact regions.
CONCLUSION
In a review published in 1991, the authors pointed out in their conclusion that AFM may prove to be a valuable tool for probing microbial surfaces at high resolution (6). Ten years later, it is fair to say that part of the dream has become reality. With AFM, the structure of isolated cell surface layers can now be imaged at (sub)molecular resolution and under aqueous solutions. Remarkably, conformational changes induced by a physiological signal can be monitored in single membrane proteins, providing novel insight into structure-function relationships. For living cells, high-resolution imaging remains challenging, but a wealth of novel structural information can be obtained, such as visualizations of surface ultrastructure in native conditions and monitoring of physiological changes in real time. Besides surface imaging, AFM force measurements can be used to probe molecular interactions, physicochemical properties, surface stiffness, and macromolecular elasticity, contributing to our knowledge of cell surface functions.
However, researchers should keep in mind that the use of AFM in microbiology remains delicate and that accurate data collection and interpretation require specific expertise. In particular, great care should be taken to optimize sample preparation procedures and imaging conditions when exploring new specimens. Also, the best results will be obtained when AFM is combined with complementary biochemical and structural techniques. In the future, it is hoped that advances in sample preparation techniques, instrumentation, and recording conditions will make it possible to observe living cells in real time with molecular resolution, monitor molecular conformational changes at cell surfaces, probe the turgor pressure in living cells, and map specific interaction sites associated with antibodies, lectins, and adhesins.