Protein targeting or protein sorting is the biological mechanism by which proteins are transported to their appropriate destinations in the cell or outside it. Proteins can be targeted to the inner space of an organelle, different intracellular membranes, plasma membrane, or to exterior of the cell via secretion. This delivery process is carried out based on information contained in the protein itself. Correct sorting is crucial for the cell; errors can lead to diseases.
Targeting signals
Targeting signals are the pieces of information that enable the cellular transport machinery to correctly position a protein inside or outside the cell. This information is contained in the polypeptide chain or in the folded protein. The continuous stretch of amino acid residues in the chain that enables targeting are called signal peptides or targeting peptides. There are two types of targeting peptides, the presequences and the internal targeting peptides. The presequences of the targeting peptide are often found at the N-terminal extension and is composed of between 6-136 basic and hydrophobic amino acids. In case of peroxisomes the targeting sequence is on the C-terminal extension mostly. Other signals, known as signal patches, are composed of parts which are separate in the primary sequence. They become functional when folding brings them together on the protein surface. In addition, protein modifications like glycosylations can induce targeting.
Protein translocation
In 1970, Günter Blobel conducted experiments on the translocation of proteins across membranes. He was awarded the 1999 Nobel prize for his findings. He discovered that many proteins have a signal sequence, that is, a short amino acid sequence at one end that functions like a postal code for the target organelle. The translation of mRNA into protein by a ribosome takes place within the cytosol. If the synthesized proteins “belong” in a different organelle, they can be transported there in either of two ways depending on the protein: Co-translational translocation (translocation during the process of translation), and post-translational translocation (translocation after the process of translation is complete).
Co-translational translocation
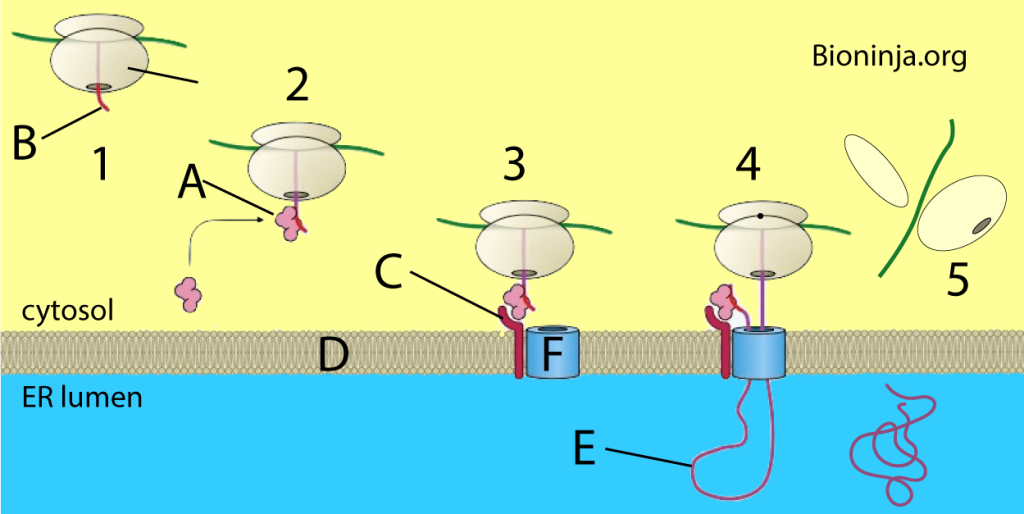
Most proteins that are secretory, membrane-bound, or reside in the endoplasmic reticulum (ER), golgi or endosomes use the co-translational translocation pathway. This process begins with the N-terminal signal peptide of the protein being recognized by a signal recognition particle (SRP) while the protein is still being synthesized on the ribosome. The synthesis pauses while the ribosome-protein complex is transferred to an SRP receptor on the ER in eukaryotes, and the plasma membrane in prokaryotes. There, the nascent protein is inserted into the translocon, a membrane-bound protein conducting channel composed of the Sec61 translocation complex in eukaryotes, and the homologous SecYEG complex in prokaryotes. In secretory proteins and type I transmembrane proteins, the signal sequence is immediately cleaved from the nascent polypeptide once it has been translocated into the membrane of the ER (eukaryotes) or plasma membrane (prokaryotes) by signal peptidase. The signal sequence of type II membrane proteins and some polytopic membrane proteins are not cleaved off and therefore are referred to as signal anchor sequences. Within the ER, the protein is first covered by a chaperone protein to protect it from the high concentration of other proteins in the ER, giving it time to fold correctly. Once folded, the protein is modified as needed (for example, by glycosylation), then transported to the Golgi for further processing and goes to its target organelles or is retained in the ER by various ER retention mechanisms.
The amino acid chain of transmembrane proteins, which often are transmembrane receptors, passes through a membrane one or several times. They are inserted into the membrane by translocation, until the process is interrupted by a stop-transfer sequence, also called a membrane anchor or signal-anchor sequence. These complex membrane proteins are at the moment mostly understood using the same model of targeting that has been developed for secretory proteins. However, many complex multi-transmembrane proteins contain structural aspects that do not fit the model. Seven transmembrane G-protein coupled receptors (which represent about 5% of the genes in humans) mostly do not have an amino-terminal signal sequence. In contrast to secretory proteins, the first transmembrane domain acts as the first signal sequence, which targets them to the ER membrane. This also results in the translocation of the amino terminus of the protein into the ER membrane lumen. This would seem to break the rule of “co-translational” translocation which has always held for mammalian proteins targeted to the ER. This has been demonstrated with opsin with in vitro experiments.A great deal of the mechanics of transmembrane topology and folding remains to be elucidated.
Post-translational translocation
Even though most secretory proteins are co-translationally translocated, some are translated in the cytosol and later transported to the ER/plasma membrane by a post-translational system. In prokaryotes this requires certain cofactors such as SecA and SecB. This pathway is poorly understood in eukaryotes, but is facilitated by Sec62 and Sec63, two membrane-bound proteins.
In addition, proteins targeted to other destinations, such as mitochondria, chloroplasts, or peroxisomes, use specialized post-translational pathways. Also, proteins targeted for the nucleus are translocated post-translation. They pass through the nuclear envelope via nuclear pores.
Comments are closed