The nested qPCR or the nested RT-PCR gives great power to this technique which can be a helpful method for the phylogenetic analysis and identification of different pathogens.
The technique has higher sensitive hence even if the sample contains lower DNA, it can amplify, which is not possible by the conventional PCR technique. In addition to this, the method is highly specific.
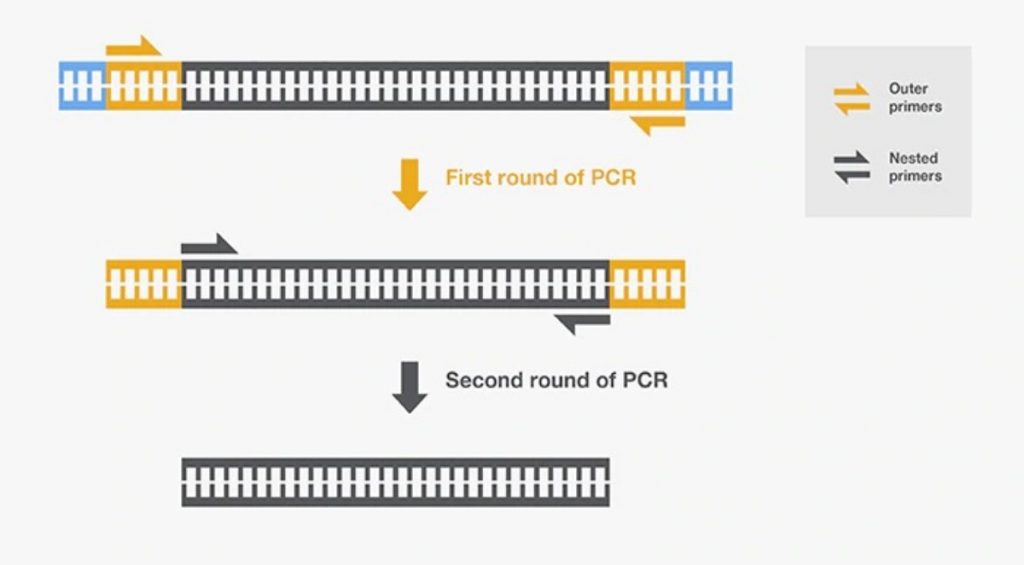
In the nested real-time PCR, the universal primers for 16S and 18S rRNA are used as an outer primer.

Once it amplifies into the PCR machine, the set of species-specific or unique sequence primers are used as an inner set of primers.
The second round of PCR or multiplex PCR (more set of primers for different species), individual PCR or the target-specific PCR technique can be applied.
The unique sequence primers are specific to one pathogen which amplifies the template DNA if the target sequence is present.
Once the amplification is achieved, the amount of pathogen present in the sample is measured quantitatively-ultimately the species of the pathogen can be identified.

By using the universal primer and sequence-specific primer phylogenetic tree for different species of the pathogen can be prepared as well.
Conclusion:
Although the nested PCR is the best choice for achieving the specificity, it consumes more time. It is restricted, the technique is not suitable for long-range PCR.
Still, the nested PCR is one of the gold standard method used in the identification of pathogens. The combined multiplex-nested PCR method is used in the study of 16s rRNA and 18s rRNA of HCV and HSV.
Comments are closed