The process of the ARMS PCR is simple and very effective. No harmful radiolabelling is involved in the ARMS allele-specific PCR.
The procedure of ARMS PCR is divided into 4 steps:
1. Primer designing
2. Amplification
3. Agarose gel electrophoresis
4. Results interpretations
1. Primer designing for ARMS PCR:
The primer must be allele-specific.
There are several points we have to understand while designing a primer for the ARMS-PCR.
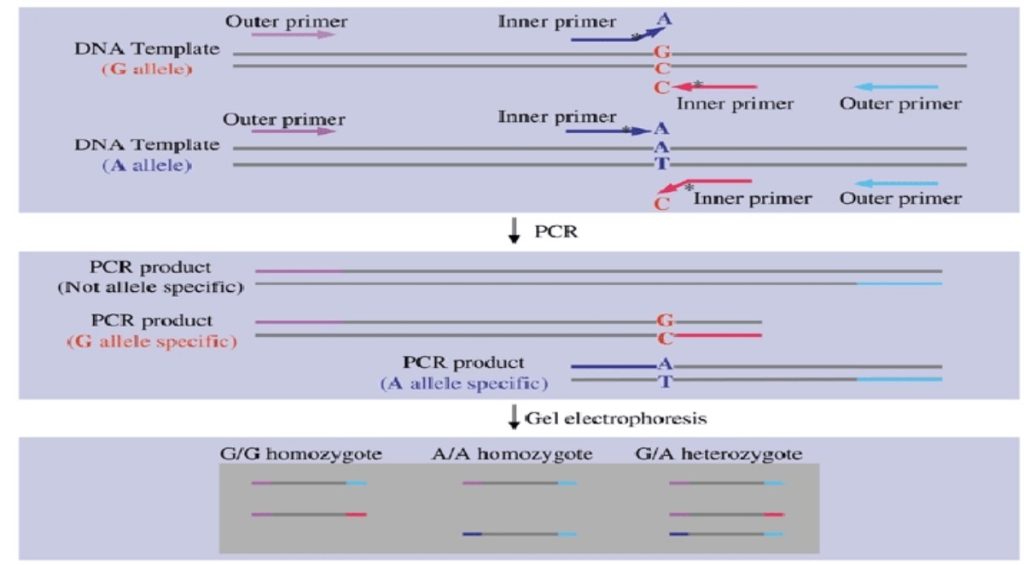
Now carefully follow the figure above,
Suppose our DNA sequence has G-A point mutation viz, G in normal allele and A in place of G in the mutant allele.
We have to design a forward primer in such a manner that for normal allele the primer contains C (complementary to G) at 3’ end and the mutant primer contains T in place of C.
The magic of the present technique has happened when we add a mismatch base near to our SNP at the 3’ end.
Why we have to add mismatch?
The mismatch is the key factor in achieving the amplification. If the mismatch is weak the chance of amplification is higher. Add strong mismatch near the 3’ end of the primer (at -2 position ideally) hence, in the non-complementary allele it can not bind to the DNA sequences and therefore terminates amplification.
C:T, G:A and A:G are the strong mismatch base pairs which reduce the amplification process up to 100 fold.
The reverse primer or another primer (which is not modified) generally remains the same. Furthermore, the primer must fulfil all the criteria for the ideal primer. Read the primer design guideline: PCR primer design guidelines
The primer length of the allele-specific primer must be between 20 to 30 nucleotides.
Once our primer is ready we have to modify the amplification conditions for achieving higher amplification.
2.Amplification conditions:
The annealing temperature should higher (do not compromise the annealing temperature).
The concentration of the PCR components is given into the table below,

The PCR reaction preparation recipe.
Importantly, the PCR cycles for ARMS-PCR are lower than the normal PCR reaction. Set a PCR cycle between 22 to 25 but not 35. As the PCR cycles increase the chance of false-positive results increases.
Here in the ARMS PCR, the internal control plays a crucial role, it facilitates additional accuracy in the allele-specific PCR by reducing the chance of the false-positive results.
3. Agarose gel electrophoresis:
The beauty of the ARMS PCR is that it does not require any hybridization steps. Which will make this technique more suitable and more reliable for the diagnosis.
The amplified fragments are directly loaded on the 2% agarose gel for getting the results. The samples are loaded sequentially on the agarose gel and run for 45 minutes.

4. Results and interpretation:
First, observe the internal control. If the internal controls bands are present in all the wells indicates that our reaction is completely fine, no false-positive results are present.
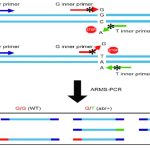
Now analyse each band in normal as well as in mutant allele. The graphical representation of the agarose gel electrophoresis results are shown in the figure below,
Suppose we have three samples one normal, one heterozygous carrier and one homozygous dominant.
We have prepared 2 tubes for each sample, a total of 6 tubes with one positive control and two negative control.
See the figure,

As shown into the figure above, an only single band is observed in the homozygous normal sample, two bands (one for normal and one for mutant) are observed in the heterozygous carrier sample and a single band of a mutant allele is observed in the homozygous disease condition. The wells 7, 8 and 9 are the controls.
Importance of ARMS-PCR in the diagnosis of genetic disease:
The ARMS PCR is one of the important tools in the genetic disease diagnosis in recent days. The restriction digestion method is not 100% accurate and also, not all the sequences have the restriction site.
Therefore, restriction digestion is not applicable in all types of mutation or polymorphism.
The allele-specific PCR is an accurate method for single-gene disorders having the SNPs. It is also a gold standard method for Sickle cell anaemia and thalassemia like inherited disorder.
Also, it is applicable in mutation detection of JAK2 and HIV. Commercial kits are now available for the different range of disorders such as HbS and JAK2 gene SNPs detection kits.
The advantage of ARMS PCR:
· The technique is exclusively for the SNP genotyping.
· Further, homozygous and heterozygous can be detected by this technique
· It is widely applicable in the single gene point mutation detection such as sickle cell anaemia and thalassemia.
· It is fast, reliable accurate and rapid.
Limitation of ARMS PCR:
· Deletion/other major duplication and chromosomal abnormalities cannot be detected.
· Only known SNPs are detected by ARMS-PCR.
· Internal control is required because of the chance of the false-negative results.
· It is temperature-sensitive. A minute fluctuation in the temperature leads to false-positive results.
· Thousands of SNPs cannot be detected in a single assay.
The high fidelity Taq DNA polymerase has the 3′ to 5′ exonuclease activity which removes the mismatch bases. that is why it is not used in the ARMS-PCR.
Conclusion:
The ARMS-PCR method is popular for the detection of known SNPs, however, it is a very tedious and time-consuming process to encounter more SNPs at once. Because it is allele-specific, the accuracy of ARMS-PCR is higher.
Some of the deletions can also be screened using ARMS-PCR method.